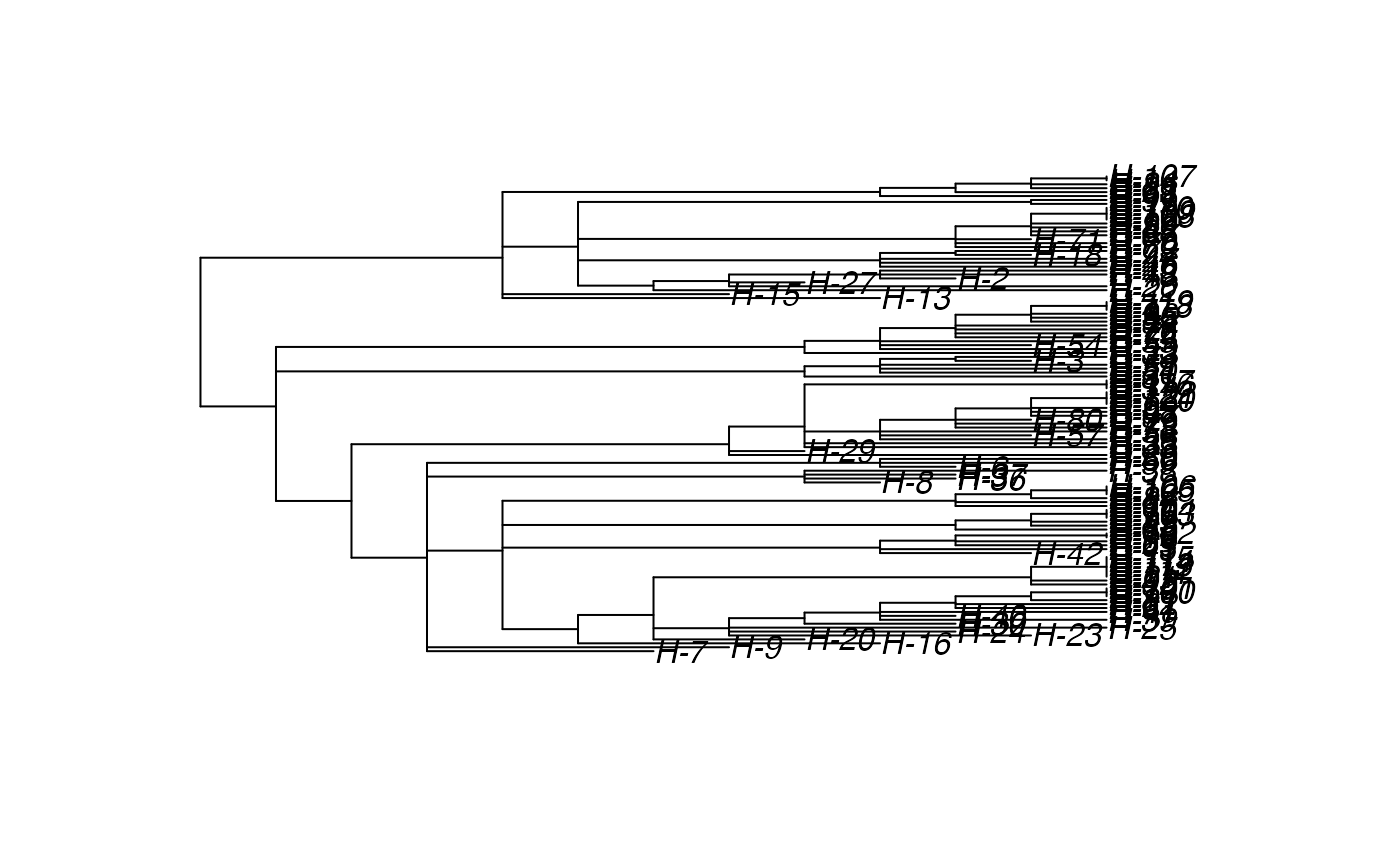
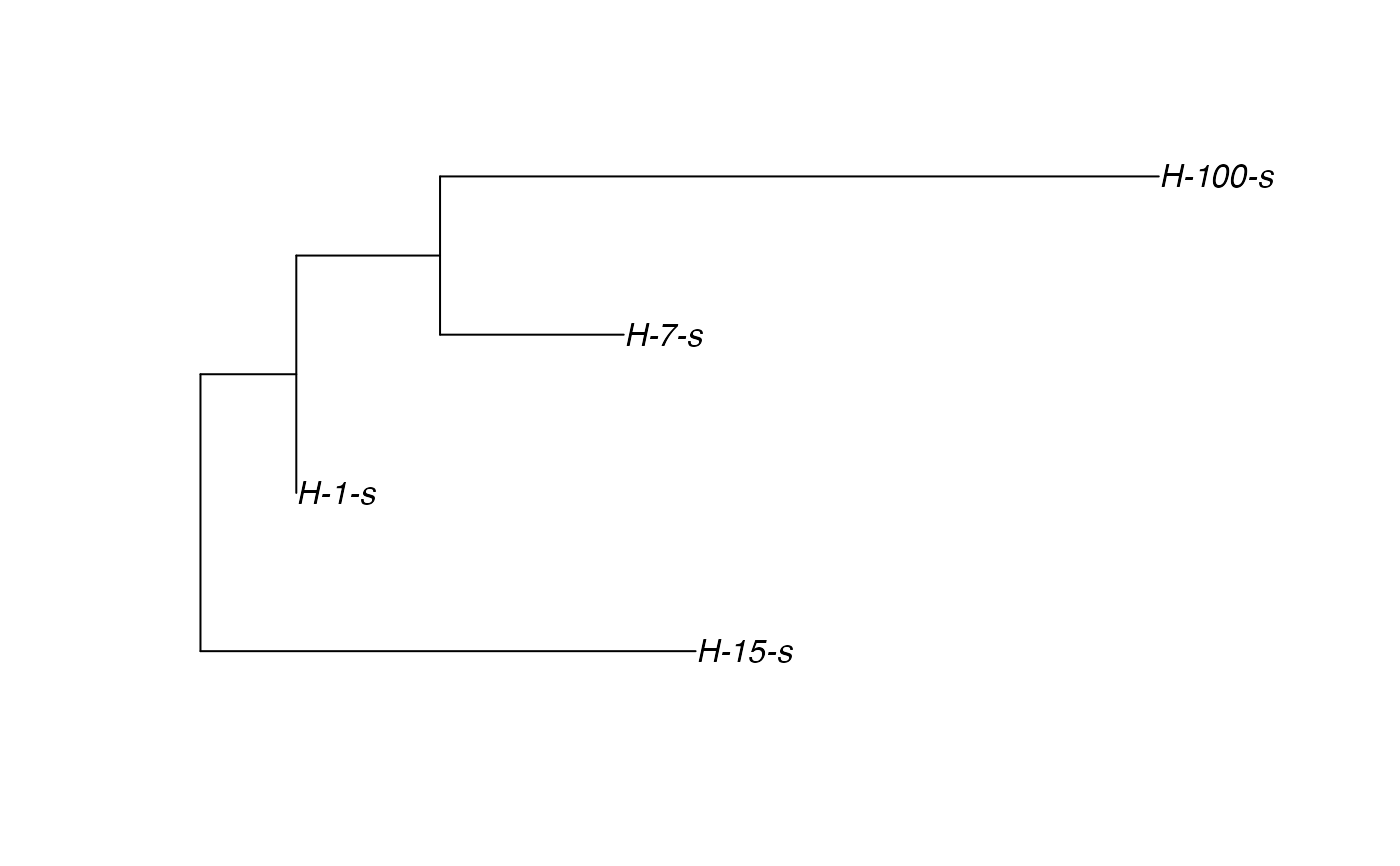
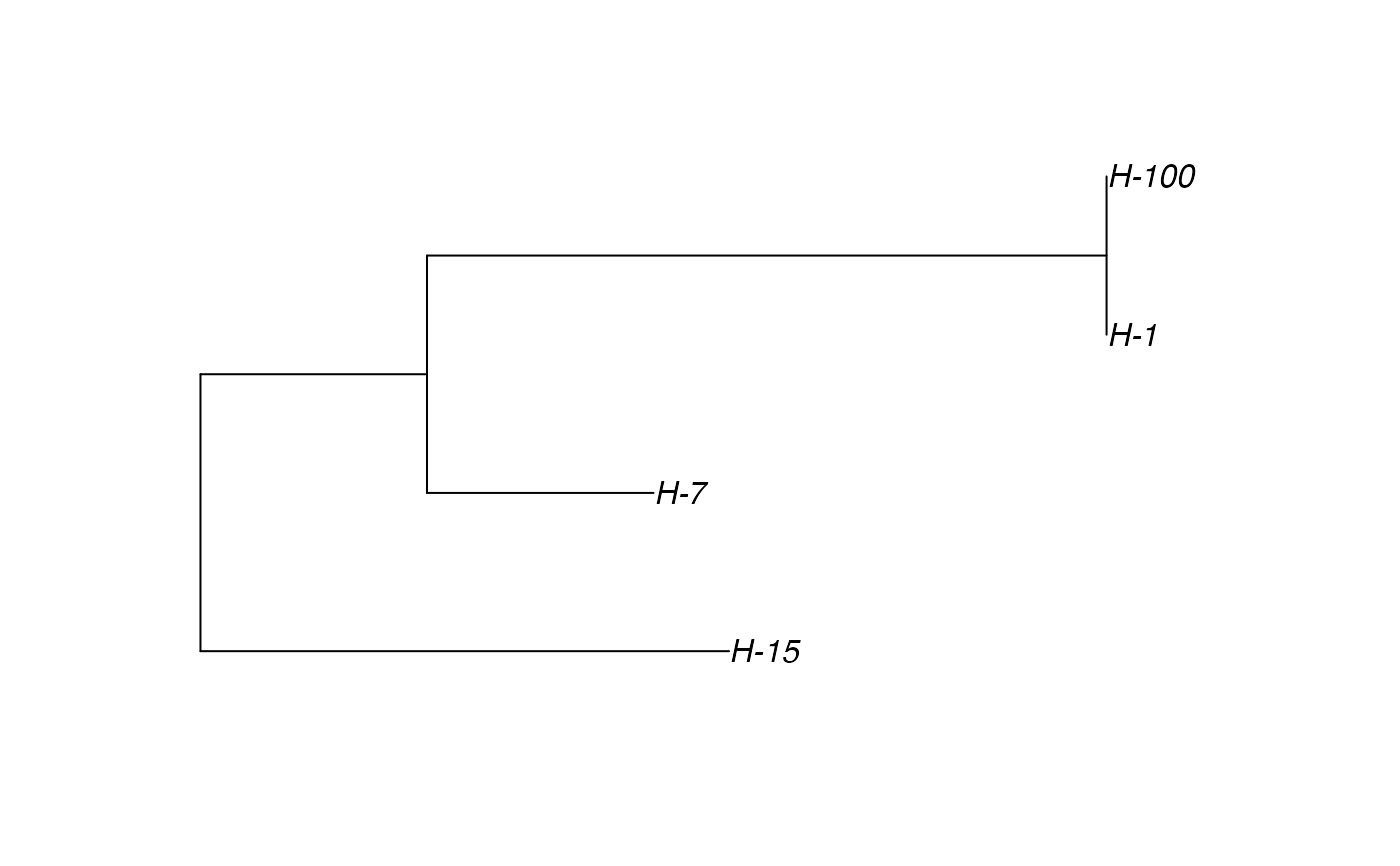
Sample the transmission tree (phylogenetic tree-like) among the exited hosts
Source:R/output_treeGenerator.r
sampleTransmissionTreeFromExiting.RdSample a full transmission tree. This function allows for sampling only exited (i.e. inactive) individuals (e.g. when the sampling procedure is destructive or cuts the hosts from the population). Beware because it does not influence the epidemiological process, it only means that the host has been sampled when exiting the simulation.
sampleTransmissionTreeFromExiting(tree, hosts)
Arguments
| tree | a |
|---|---|
| hosts | a vector of dead hosts to sample |
Value
A tree of class treedata, containing a
phylogenetic tree based on the transmission chain and the mapped data at all the nodes.
Details
The tree needs to be produced by function getTransmissionTree
applied on the same nosoiSim object.
See also
For exporting the annotated tree to other software packages, see functions
in treeio (e.g. write.beast).
To get the full transmission matrix, see getTransmissionTree.
For sampling among non-dead individuals, see sampleTransmissionTree.
Examples
# \donttest{ t_incub_fct <- function(x){rnorm(x,mean = 5,sd=1)} p_max_fct <- function(x){rbeta(x,shape1 = 5,shape2=2)} p_Exit_fct <- function(t){return(0.08)} p_Move_fct <- function(t){return(0.1)} proba <- function(t,p_max,t_incub){ if(t <= t_incub){p=0} if(t >= t_incub){p=p_max} return(p) } time_contact = function(t){round(rnorm(1, 3, 1), 0)} transition.matrix = matrix(c(0, 0.2, 0.4, 0.5, 0, 0.6, 0.5, 0.8, 0), nrow = 3, ncol = 3, dimnames = list(c("A", "B", "C"), c("A", "B", "C"))) set.seed(805) test.nosoi <- nosoiSim(type="single", popStructure="discrete", length=20, max.infected=100, init.individuals=1, init.structure="A", structure.matrix=transition.matrix, pMove=p_Move_fct, param.pMove=NA, nContact=time_contact, param.nContact=NA, pTrans = proba, param.pTrans = list(p_max=p_max_fct, t_incub=t_incub_fct), pExit=p_Exit_fct, param.pExit=NA )#> #>#>#>#> #>## Make sure all needed packages are here if (requireNamespace("ape", quietly = TRUE) || requireNamespace("tidytree", quietly = TRUE) || requireNamespace("treeio", quietly = TRUE)) { library(ape) library(tidytree) library(treeio) #' ## Full transmission tree ttreedata <- getTransmissionTree(test.nosoi) plot(ttreedata@phylo) ## Sampling "non dead" individuals hID <- c("H-1", "H-7", "H-15", "H-100") samples <- data.table(hosts = hID, times = c(5.2, 9.3, 10.2, 16), labels = paste0(hID, "-s")) sampledTree <- sampleTransmissionTree(test.nosoi, ttreedata, samples) plot(sampledTree@phylo) ## Sampling "dead" individuals sampledDeadTree <- sampleTransmissionTreeFromExiting(ttreedata, hID) plot(sampledDeadTree@phylo) }# }