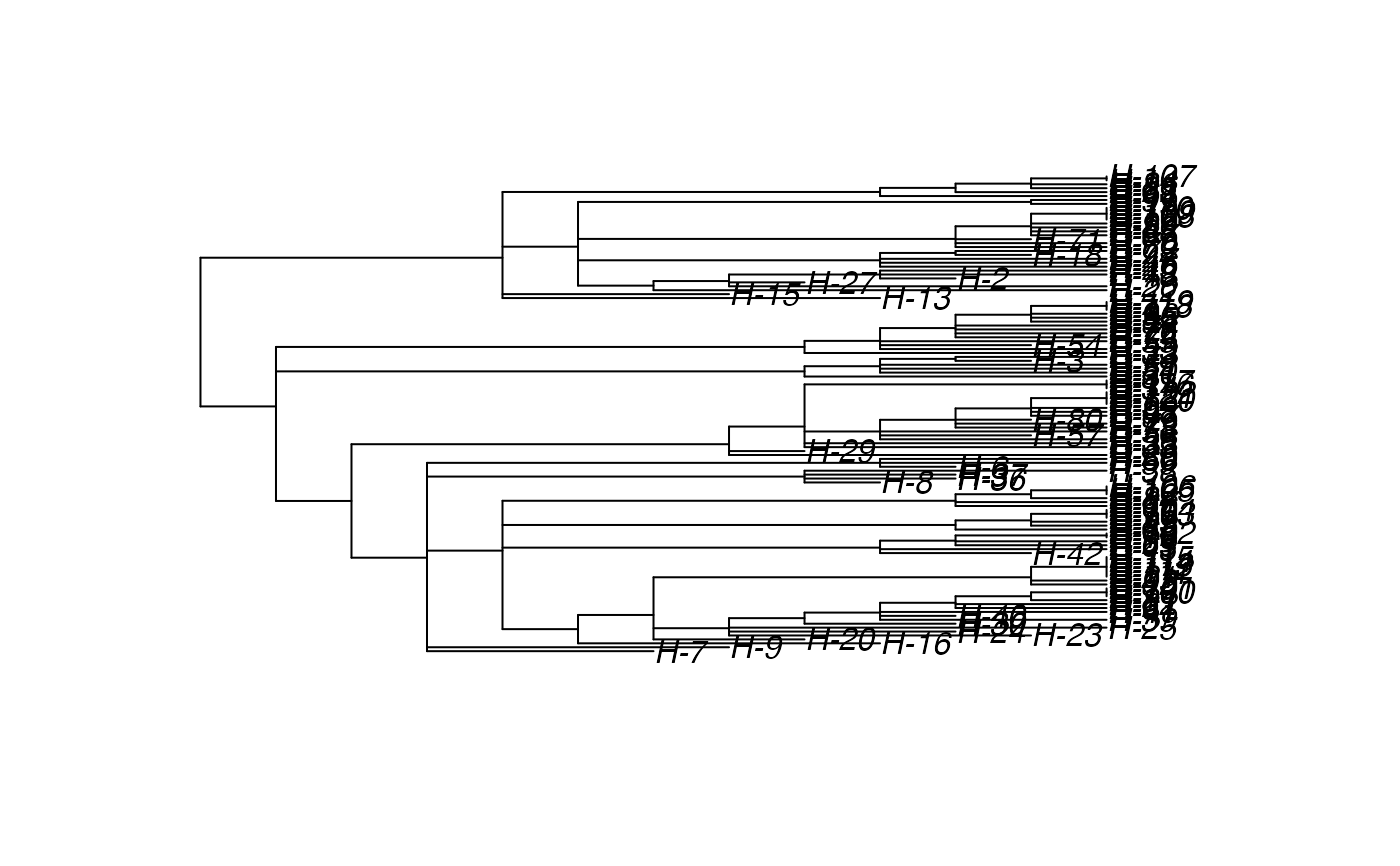
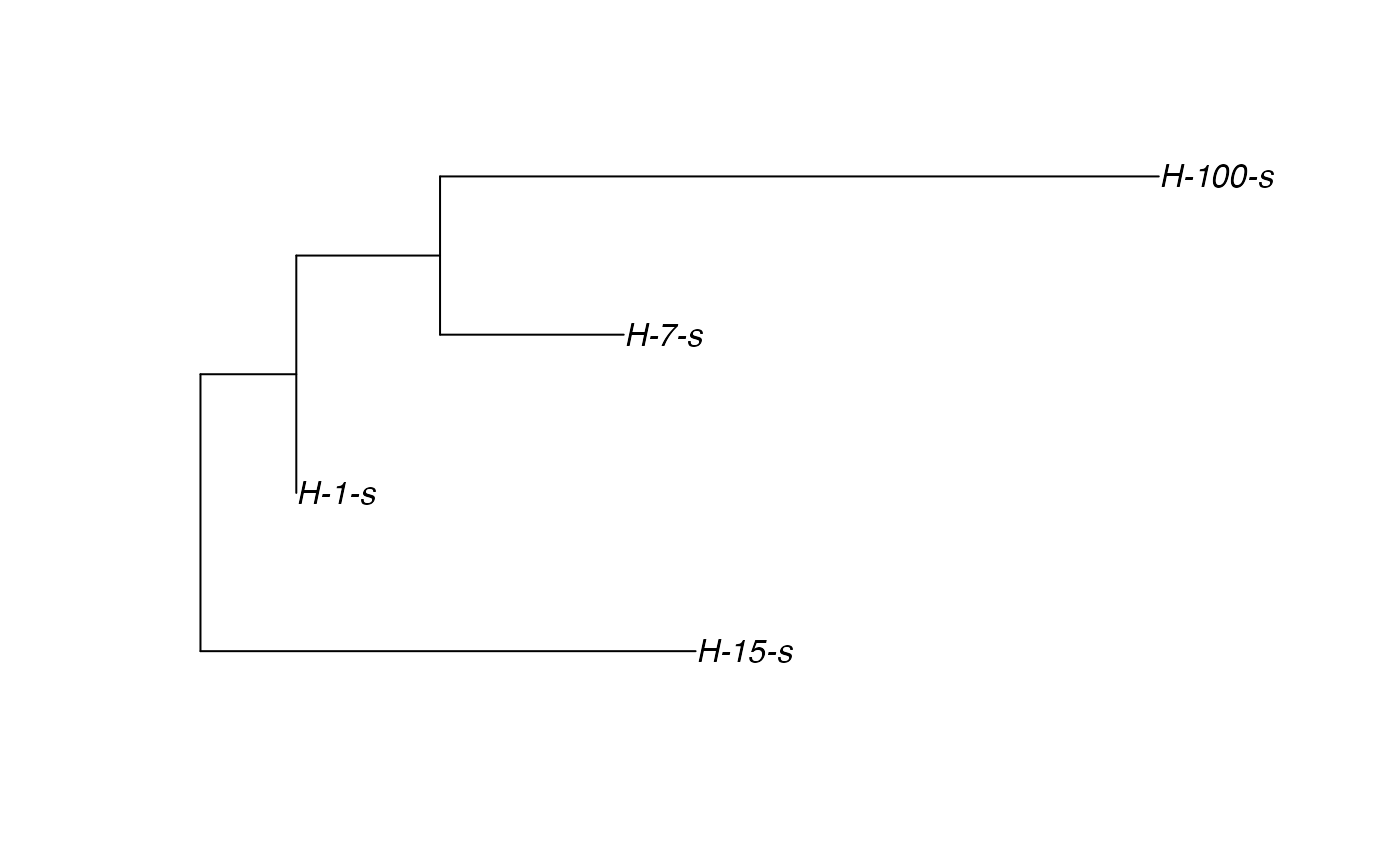
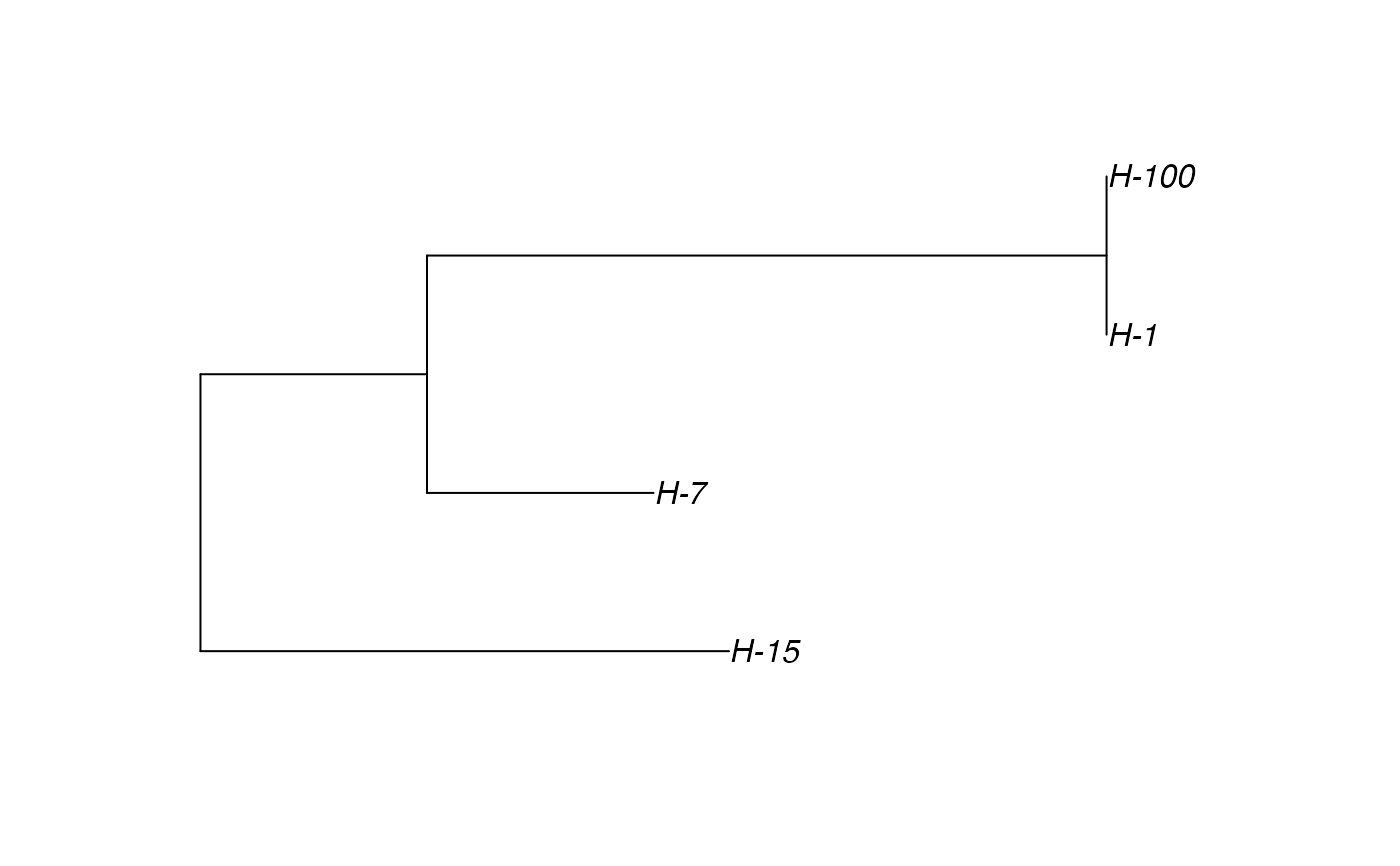
Gets the full transmission tree (phylogenetic tree-like) from a nosoi simulation
Source: R/output_treeGenerator.r
getTransmissionTree.RdFrom a nosoi simulated epidemics, this function extracts the full transmission tree in a form mimicking a phylogenetic tree.
getTransmissionTree(nosoiInf)
Arguments
| nosoiInf | an object of class |
|---|
Value
A tree of class treedata, containing a
phylogenetic tree based on the transmission chain and the mapped data at all the nodes.
Details
This function uses packages tidytree and treeio,
that rely on ape.
See also
For exporting the annotated tree to other software packages, see functions
in treeio (e.g. write.beast).
To sub-sample this tree, see functions sampleTransmissionTree and sampleTransmissionTreeFromExiting
Examples
# \donttest{ t_incub_fct <- function(x){rnorm(x,mean = 5,sd=1)} p_max_fct <- function(x){rbeta(x,shape1 = 5,shape2=2)} p_Exit_fct <- function(t){return(0.08)} p_Move_fct <- function(t){return(0.1)} proba <- function(t,p_max,t_incub){ if(t <= t_incub){p=0} if(t >= t_incub){p=p_max} return(p) } time_contact = function(t){round(rnorm(1, 3, 1), 0)} transition.matrix = matrix(c(0, 0.2, 0.4, 0.5, 0, 0.6, 0.5, 0.8, 0), nrow = 3, ncol = 3, dimnames = list(c("A", "B", "C"), c("A", "B", "C"))) set.seed(805) test.nosoi <- nosoiSim(type="single", popStructure="discrete", length=20, max.infected=100, init.individuals=1, init.structure="A", structure.matrix=transition.matrix, pMove=p_Move_fct, param.pMove=NA, nContact=time_contact, param.nContact=NA, pTrans = proba, param.pTrans = list(p_max=p_max_fct, t_incub=t_incub_fct), pExit=p_Exit_fct, param.pExit=NA )#> #>#>#>#> #>## Make sure all needed packages are here if (requireNamespace("ape", quietly = TRUE) || requireNamespace("tidytree", quietly = TRUE) || requireNamespace("treeio", quietly = TRUE)) { library(ape) library(tidytree) library(treeio) #' ## Full transmission tree ttreedata <- getTransmissionTree(test.nosoi) plot(ttreedata@phylo) ## Sampling "non dead" individuals hID <- c("H-1", "H-7", "H-15", "H-100") samples <- data.table(hosts = hID, times = c(5.2, 9.3, 10.2, 16), labels = paste0(hID, "-s")) sampledTree <- sampleTransmissionTree(test.nosoi, ttreedata, samples) plot(sampledTree@phylo) ## Sampling "dead" individuals sampledDeadTree <- sampleTransmissionTreeFromExiting(ttreedata, hID) plot(sampledDeadTree@phylo) }#> #>#> #> #>#> #>#> #> #>#> #> #>#> #> #>#> #> #> #> #>#> #>#> #> #>#> #> #>#> Warning: `mutate_()` was deprecated in dplyr 0.7.0. #> Please use `mutate()` instead. #> See vignette('programming') for more help# }