Sample the transmission tree (phylogenetic tree-like)
Source:R/output_treeGenerator.r
sampleTransmissionTree.RdSample a full transmission tree. This function allows for sampling multiple times on the same lineage. When this happens, the sampled ancestor is a tip with length zero.
sampleTransmissionTree(nosoiInf, tree, samples)
Arguments
| nosoiInf | an object of class |
|---|---|
| tree | a |
| samples | a
|
Value
A tree of class treedata, containing a
phylogenetic tree based on the transmission chain and the mapped data at all the nodes.
Details
The tree needs to be produced by function getTransmissionTree
applied on the same nosoiSim object.
See also
For exporting the annotated tree to other software packages, see functions
in treeio (e.g. write.beast).
To get the full transmission matrix, see getTransmissionTree.
For sampling only dead individuals, see sampleTransmissionTreeFromExiting.
Examples
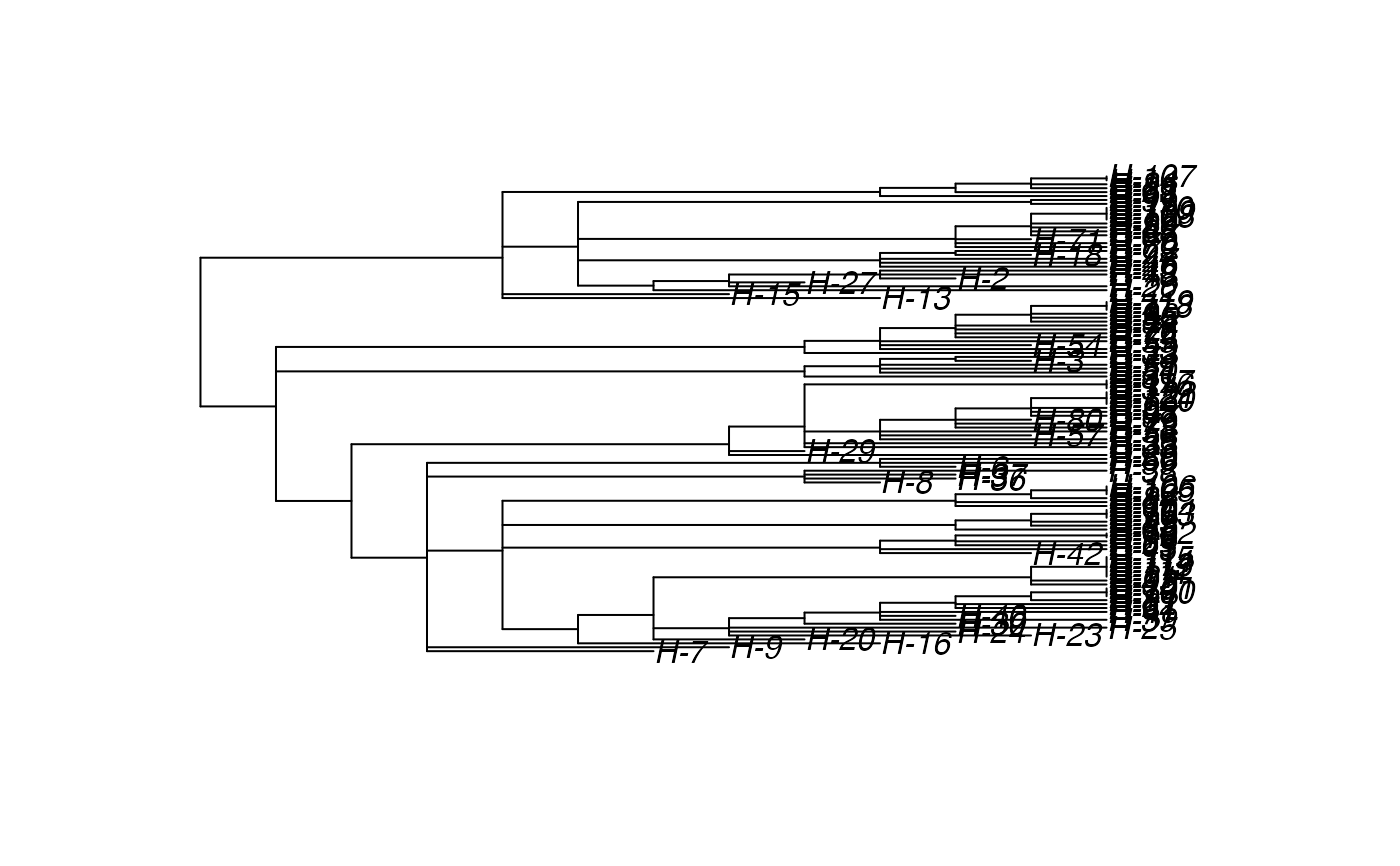
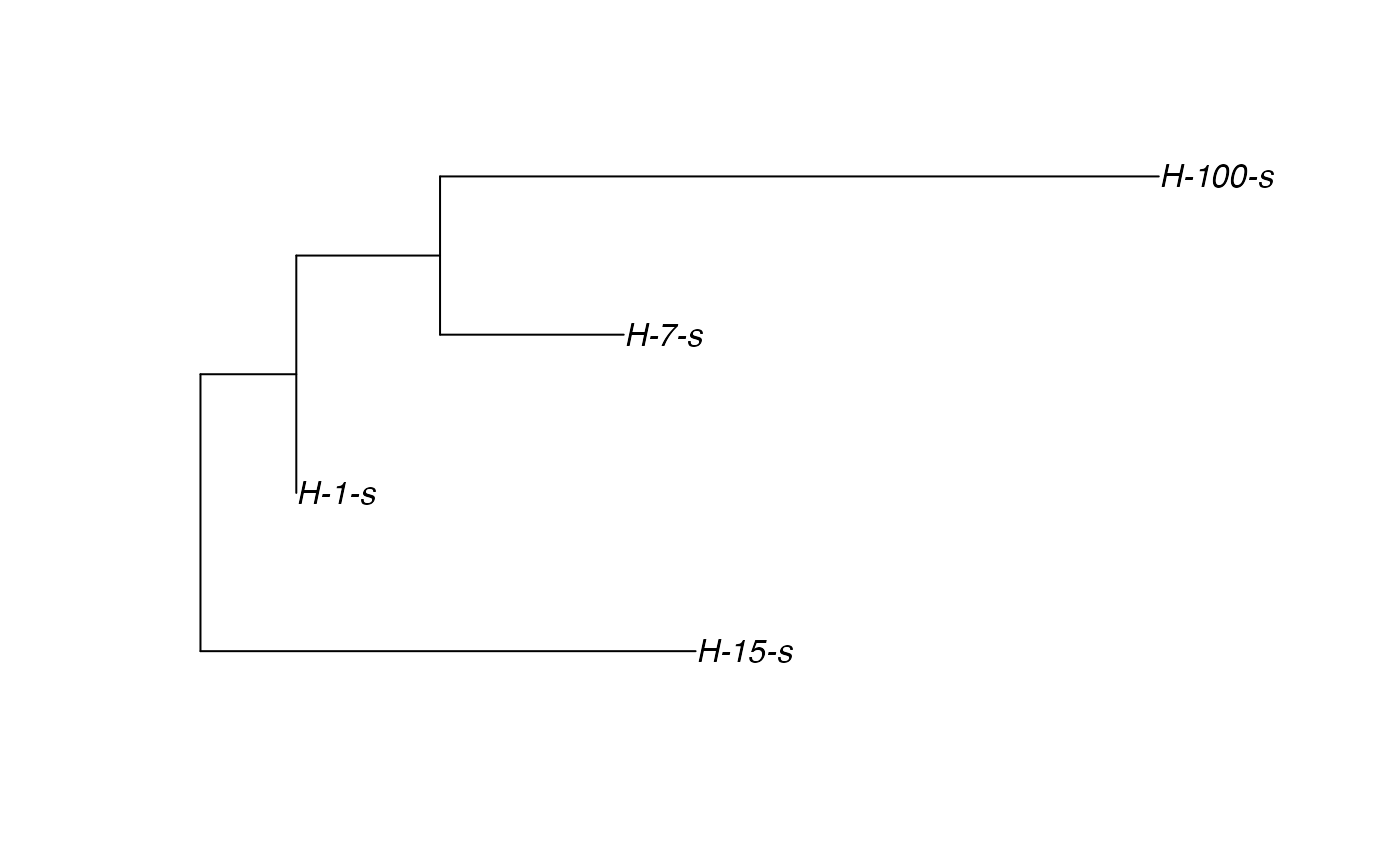
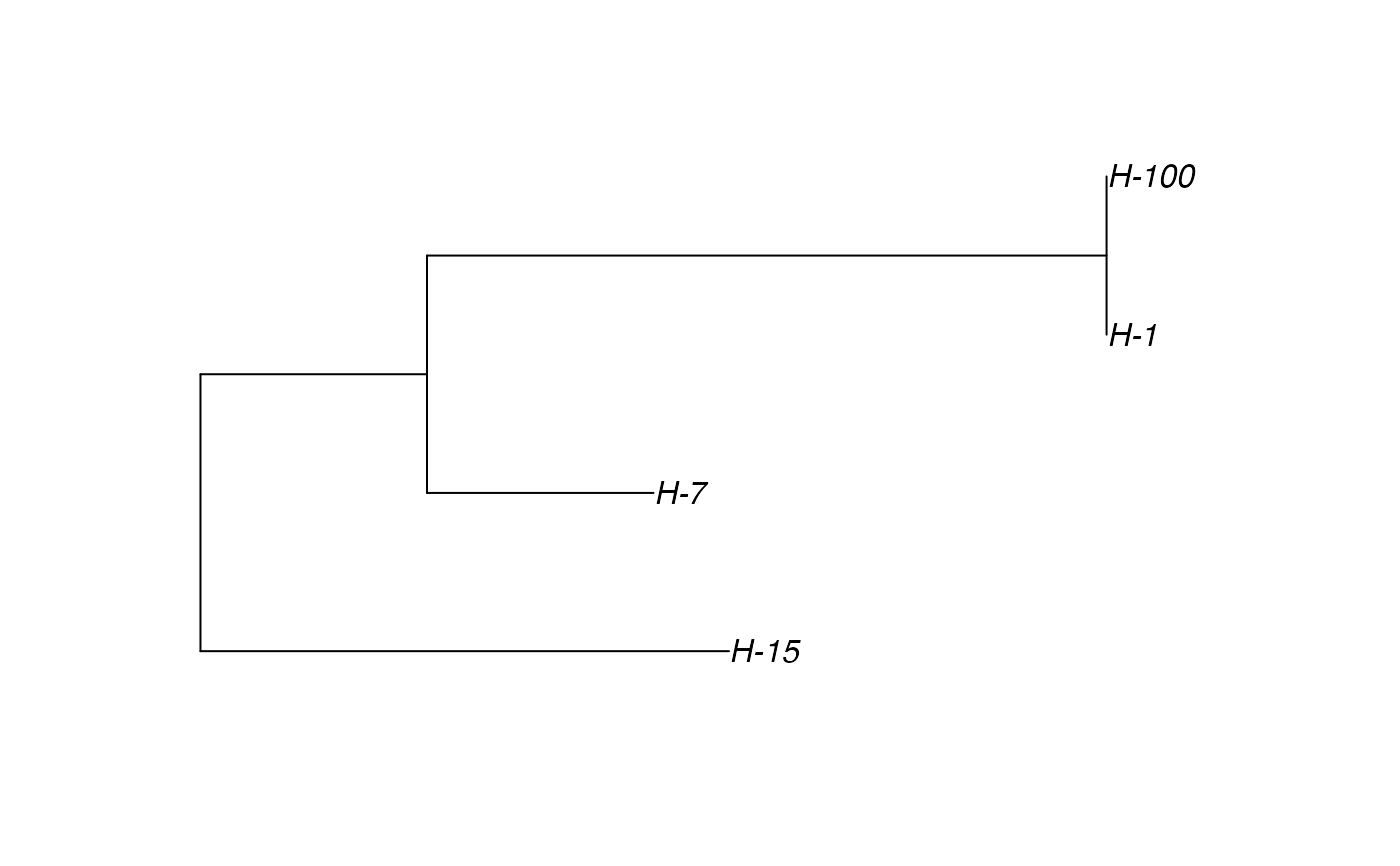
# \donttest{ t_incub_fct <- function(x){rnorm(x,mean = 5,sd=1)} p_max_fct <- function(x){rbeta(x,shape1 = 5,shape2=2)} p_Exit_fct <- function(t){return(0.08)} p_Move_fct <- function(t){return(0.1)} proba <- function(t,p_max,t_incub){ if(t <= t_incub){p=0} if(t >= t_incub){p=p_max} return(p) } time_contact = function(t){round(rnorm(1, 3, 1), 0)} transition.matrix = matrix(c(0, 0.2, 0.4, 0.5, 0, 0.6, 0.5, 0.8, 0), nrow = 3, ncol = 3, dimnames = list(c("A", "B", "C"), c("A", "B", "C"))) set.seed(805) test.nosoi <- nosoiSim(type="single", popStructure="discrete", length=20, max.infected=100, init.individuals=1, init.structure="A", structure.matrix=transition.matrix, pMove=p_Move_fct, param.pMove=NA, nContact=time_contact, param.nContact=NA, pTrans = proba, param.pTrans = list(p_max=p_max_fct, t_incub=t_incub_fct), pExit=p_Exit_fct, param.pExit=NA )#> #>#>#>#> #>## Make sure all needed packages are here if (requireNamespace("ape", quietly = TRUE) || requireNamespace("tidytree", quietly = TRUE) || requireNamespace("treeio", quietly = TRUE)) { library(ape) library(tidytree) library(treeio) #' ## Full transmission tree ttreedata <- getTransmissionTree(test.nosoi) plot(ttreedata@phylo) ## Sampling "non dead" individuals hID <- c("H-1", "H-7", "H-15", "H-100") samples <- data.table(hosts = hID, times = c(5.2, 9.3, 10.2, 16), labels = paste0(hID, "-s")) sampledTree <- sampleTransmissionTree(test.nosoi, ttreedata, samples) plot(sampledTree@phylo) ## Sampling "dead" individuals sampledDeadTree <- sampleTransmissionTreeFromExiting(ttreedata, hID) plot(sampledDeadTree@phylo) }# }